Neural Jastrow#
In this tutorial, we use neural networks as generalized Jastrow factors to study the Heisenberg J1-J2 model.
Reference:
import numpy as np
import matplotlib.pyplot as plt
import jax.numpy as jnp
import equinox as eqx
import quantax as qtx
from IPython.display import clear_output
lattice = qtx.sites.Square(4, Nparticles=(8, 8))
N = lattice.Nsites
The Hamiltonian is
where \(\braket{ij}\) and \(\braket{\braket{ij}}\) represent nearest and next-nearest neighbors, respectively.
from quantax.symmetry import Identity, TransND, C4v, SpinInverse
H = qtx.operator.Heisenberg(J=[1, 0.5], n_neighbor=[1, 2])
full_symm = TransND() @ C4v() @ SpinInverse()
E, psi = H.diagonalize(symm=full_symm)
exact_energy = E[0]
exact_state = qtx.state.DenseState(psi, full_symm)
print("Exact ground state energy:", exact_energy)
Exact ground state energy: -33.83169340557937
/home/aochen/quantax_env/lib/python3.12/site-packages/quantax/symmetry/symmetry.py:288: GeneralBasisWarning: using non-commuting symmetries can lead to unwanted behaviour of general basis, make sure that quantum numbers are invariant under non-commuting symmetries!
basis = spin_basis_general(
Gutzwiller-projected fermionic wavefunction#
The Hamiltonian in spin degrees of freedom can be mapped to fermion ones by
where \(\alpha = x,y,z\) and \(\sigma^\alpha\) are Pauli matrices. Quantax utilizes this relation internally such that the mean-field state can be optimized in spin systems.
A fermionic wavefunction \(\ket{\psi_0}\) contains redundancy of zero occupancy and double occupancy when it is used for spin systems. Therefore, one needs a Gutzwiller projection to obtain
where \(\hat P_G = \prod_i (n_{i\uparrow} - n_{i\downarrow})^2\) projects the state to spin degrees of freedom. In variational Monte Carlo, this is done by sampling suitable sectors instead of modifying the mean-field state.
Here we show how to optimize the Gutzwiller-projected fermionic mean-field wavefunction. GeneralPfState is a subclass of Variational, so one can train it directly with SR.
state = qtx.state.GeneralPfState(max_parallel=32768)
sampler = qtx.sampler.SpinExchange(state, 1024, n_neighbor=[1, 2])
optimizer = qtx.optimizer.SR(state, H)
energy = qtx.utils.DataTracer()
VarE = qtx.utils.DataTracer()
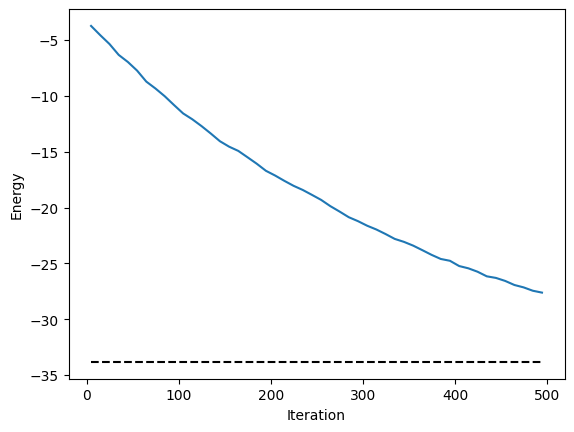
for i in range(500):
samples = sampler.sweep()
step = optimizer.get_step(samples)
state.update(step * 1e-3)
energy.append(optimizer.energy)
VarE.append(optimizer.VarE)
energy.plot(batch=10, baseline=exact_energy)
plt.xlabel("Iteration")
plt.ylabel("Energy")
plt.show()
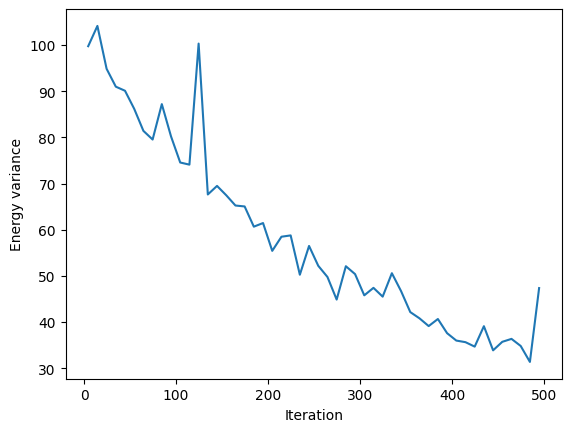
VarE.plot(batch=10)
plt.xlabel("Iteration")
plt.ylabel("Energy variance")
plt.show()
Neural Jastrow factor#
The neural Jastrow factor modifies an existing wavefunction by utilizing a neural network. The new wavefunction is given by
where \(J(s)\) is the neural Jastrow factor, and \(\psi_G(s)\) is the original Gutzwiller-projected fermionic wavefunction.
We can take the pre-trained Pfaffian state as a good initialization.
pf_model = state.model
We still utilize ResConv as the neural network. trans_symm=Identity() means network output is not summed in the last layer, so the output is a matrix \(\mathbf{J}(s)\). Then we define NeuralJastrow with trans_symm=TransND(), which means the output is
where \(T_{ij}s\) is a translation of the original input \(s\). One can view this formula as a translation symmetrized form of \(\psi(s) = J(s) \psi_G(s)\).
net = qtx.model.ResConv(nblocks=2, channels=8, kernel_size=3, trans_symm=Identity())
model = qtx.model.NeuralJastrow(net, pf_model, trans_symm=TransND())
Then one can perform the usual SR optimization on this wavefunction.
state = qtx.state.Variational(model, max_parallel=16384)
sampler = qtx.sampler.SpinExchange(state, 1024, n_neighbor=[1, 2])
optimizer = qtx.optimizer.SR(state, H)
energy = qtx.utils.DataTracer()
VarE = qtx.utils.DataTracer()
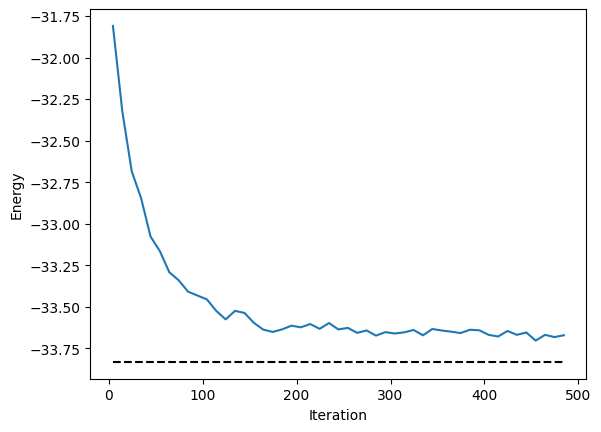
for i in range(500):
samples = sampler.sweep()
step = optimizer.get_step(samples)
state.update(step * 2e-3)
energy.append(optimizer.energy)
VarE.append(optimizer.VarE)
if i % 10 == 0:
clear_output()
energy.plot(batch=10, baseline=exact_energy)
plt.xlabel("Iteration")
plt.ylabel("Energy")
plt.show()
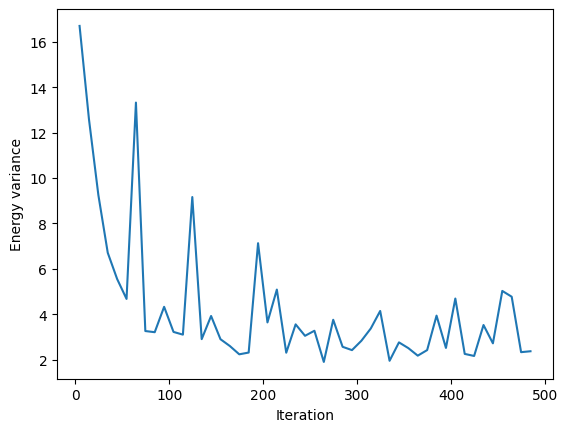
VarE.plot(batch=10)
plt.xlabel("Iteration")
plt.ylabel("Energy variance")
plt.show()
state.save("/tmp/neural_jastrow.eqx")
This state can also be symmetrized to achieve better accuracy
symm = C4v() @ SpinInverse()
state = qtx.state.Variational(
model, param_file="/tmp/neural_jastrow.eqx", symm=symm, max_parallel=2048
)
sampler = qtx.sampler.SpinExchange(state, 1024, n_neighbor=[1, 2])
optimizer = qtx.optimizer.SR(state, H)
energy = qtx.utils.DataTracer()
VarE = qtx.utils.DataTracer()
for i in range(200):
samples = sampler.sweep()
step = optimizer.get_step(samples)
state.update(step * 2e-3)
energy.append(optimizer.energy)
VarE.append(optimizer.VarE)
if i % 10 == 0:
clear_output()
energy.plot(batch=10, baseline=exact_energy)
plt.xlabel("Iteration")
plt.ylabel("Energy")
plt.show()
VarE.plot(batch=10)
plt.xlabel("Iteration")
plt.ylabel("Energy variance")
plt.show()
state.save("/tmp/neural_jastrow.eqx")
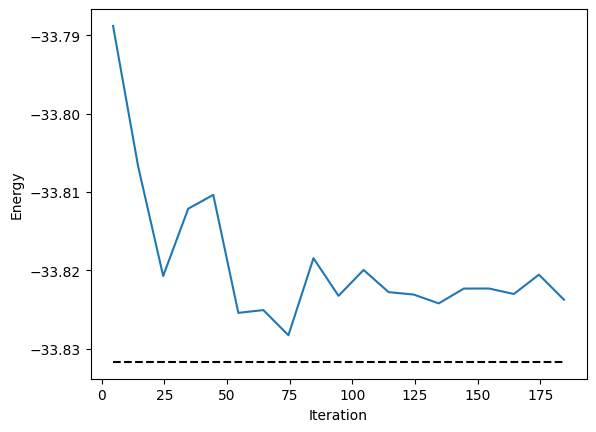
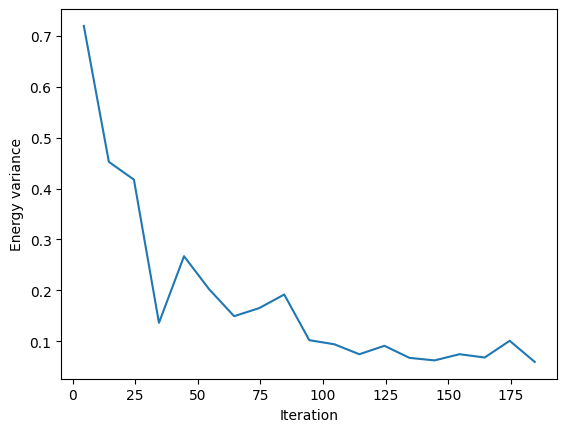
Then we can check its accuracy against the exact ground state.
dense = state.todense(full_symm).normalize()
E = dense @ H @ dense
print("Relative energy error:", abs(E - exact_energy) / abs(exact_energy))
fidelity = abs(dense @ exact_state) ** 2
print("Fidelity with exact ground state:", fidelity)
Relative energy error: 0.00017402258135205257
Fidelity with exact ground state: 0.99964914257028